A Bayesian sequential learning framework to parameterise continuum models of melanoma invasion into human skin
AP Browning, P Haridas, MJ Simpson
Bulletin of Mathematical Biology (2019)
AP Browning, P Haridas, MJ Simpson
Bulletin of Mathematical Biology (2019)
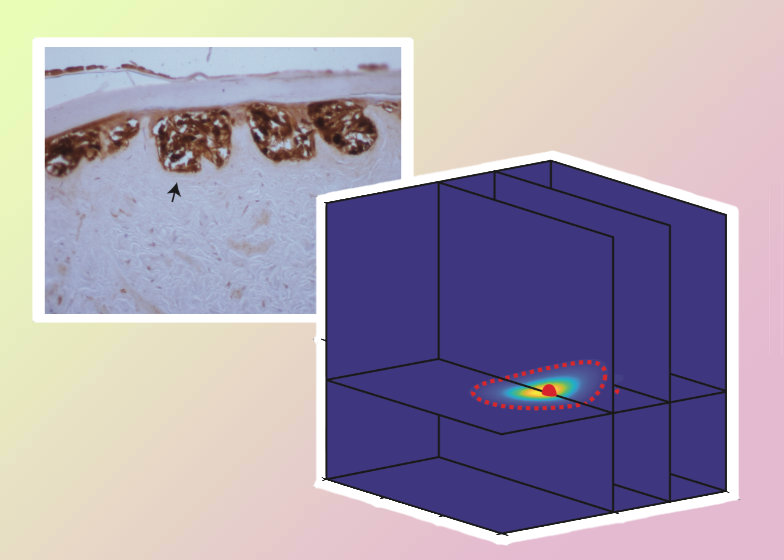
We present a novel framework to parameterise a mathematical model of cell invasion that describes how a population of melanoma cells invades into human skin tissue. Using simple experimental data extracted from complex experimental images, we estimate three model parameters: (i) the melanoma cell proliferation rate, λ; (ii) the melanoma cell diffusivity, D; and (iii) δ, a constant that determines the rate that melanoma cells degrade the skin tissue. The Bayesian sequential learning framework involves a sequence of increasingly-sophisticated experimental data from: (i) a spatially uniform cell proliferation assay; (ii) a two-dimensional circular barrier assay; and, (iii) a three-dimensional invasion assay. The Bayesian sequential learning approach leads to well-defined parameter estimates. In contrast, taking a naive approach that attempts to estimate all parameters from a single set of images from the same experiment fails to produce meaningful results. Overall our approach to inference is simple-to-implement, computationally efficient, and well-suited for many cell biology phenomena that can be described by low dimensional continuum models using ordinary differential equations and partial differential equations. We anticipate that this Bayesian sequential learning framework will be relevant in other biological contexts where it is challenging to extract detailed, quantitative biological measurements from experimental images and so we must rely on using relatively simple measurements from complex images.