Parameter identifiability and model selection for sigmoid population growth models
MJ Simpson, AP Browning, DJ Warne, OJ Maclaren, RE Baker
Journal of Theoretical Biology (2022)
MJ Simpson, AP Browning, DJ Warne, OJ Maclaren, RE Baker
Journal of Theoretical Biology (2022)
Sigmoid growth models, such as the logistic and Gompertz growth models, are widely used to study various population dynamics ranging from microscopic populations of cancer cells, to continental-scale human populations. Fundamental questions about model selection and precise parameter estimation are critical if these models are to be used to make useful inferences about underlying ecological mechanisms. However, the question of parameter identifiability for these models – whether a data set contains sufficient information to give unique or sufficiently precise parameter estimates for the given model – is often overlooked;
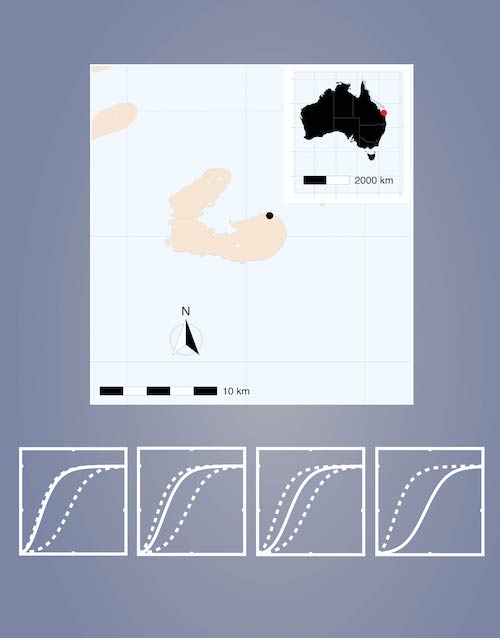
We use a profile-likelihood approach to systematically explore practical parameter identifiability using data describing the re-growth of hard coral cover on a coral reef after some ecological disturbance. The relationship between parameter identifiability and checks of model misspecification is also explored. We work with three standard choices of sigmoid growth models, namely the logistic, Gompertz, and Richards’ growth models;
We find that the logistic growth model does not suffer identifiability issues for the type of data we consider whereas the Gompertz and Richards’ models encounter practical non-identifiability issues, even with relatively-extensive data where we observe the full shape of the sigmoid growth curve. Identifiability issues with the Gompertz model lead us to consider a further model calibration exercise in which we fix the initial density to its observed value, neglecting its uncertainty. This is a common practice, but the results of this exercise suggest that parameter estimates and fundamental statistical assumptions are extremely sensitive under these conditions;
Different sigmoid growth models are used within subdisciplines within the biology and ecology literature with-out necessarily considering whether parameters are identifiable or checking statistical assumptions underlying model family adequacy. Standard practices that do not consider parameter identifiability can lead to unreliable or imprecise parameter estimates and hence potentially misleading interpretations of the underlying mechanisms of interest. While tools in this work focus on three standard sigmoid growth models and one particular data set, our theoretical developments are applicable to any sigmoid growth model and any relevant data set. MATLAB implementations of all software available on GitHub.